Isifo esibi kakhulu sokuphefumula esaziwa nge-COVID-19 - ngenxa yegciwane le-SARS-CoV-2 - saziwa ukuthi sisakazeka ngamaconsi okuphefumula kanye nokuxhumana nabantu abaseduze.[1]Umthwalo we-COVID-19 wawunzima kakhulu e-Lombardy nase-Po Valley (eNyakatho ne-Italy),[2] indawo ebonakala ngokugcwala okukhulu kwezinhlayiya, osekwaziwa kakade ukuthi zikhiqiza imiphumela engemihle empilweni yabantu.[3]Izibalo zesifunda ezitholakala e-Italy ngosuku lwango-Ephreli 12 zibonisa ukuthi cishe ama-30% abantu abanethemba njengamanje basahlala eLombardy (cishe ama-40% uma kucutshungulwa amacala aqinisekisiwe kusukela ekuqaleni kwalolu bhubhane), kulandele u-Emilia Romagna (13.5%). , Piedmont (10.5%), kanye neVeneto (10%).[2]Lezi zifunda ezine ze-Po Valley zenza ama-80% engqikithi yokufa kwabantu abaqoshwe e-Italy kanye nama-65% abamukelwa eZikhungweni Zokunakekelwa Kakhulu.[2]
Ucwaningo olwenziwa yi-Harvard School of Public Health lubonakala luqinisekisa ukuhlobana phakathi kokwenyuka kokugxila kwe-PM namazinga okufa kwabantu ngenxa ye-COVID-19 e-US[4] Kwezokuxhumana ezedlule, sicabange ukuthi kungenzeka ukuthi i-SARS-CoV-2 igciwane lingaba khona ku-particle (PM) ngesikhathi sokusabalala kokutheleleka,[5,6] ngokuhambisana nobufakazi obuvele bukhona.
itholakalela amanye amagciwane.[7-15] Nokho, udaba lwe-microbiome ehambisana ne-PM emoyeni, ikakhulukazi ezindaweni zasemadolobheni, lusaphenywa kakhulu,[16] futhi - okwamanje - akekho osaqhube izifundo zokuhlola ezihloselwe ngokuqondile. ekuqinisekiseni noma ngaphandle kokuba khona kwe-SARS-CoV-2 ku-PM.
Lapha, sethula imiphumela yokuqala yokuhlaziya esikwenzile kumasampula angu-34 PM10 e-PM10 yangaphandle/esemoyeni evela endaweni yezimboni yesiFundazwe saseBergamo, eqoqwe namasampula omoya amabili ahlukene esikhathini esiqhubekayo samasonto amathathu, kusukela ngomhlaka-21 February kuya ku-March. 13 kwe.
Ukulandela indlela echazwe nguPan et al.ngo-2019 (ekuqoqweni, ukulinganiswa kwezinhlayiyana kanye nokutholwa kwamagciwane asemoyeni), [17] amasampula e-PM aqoqwe kuzihlungi ze-quartz fiber kusetshenziswa isampuli yomoya ye-gravimetric ephansi ye-gravimetric (38.3 l/min ngamahora angama-23), ngokuhambisana nendlela yereferensi EN12341 :2014 yokuqapha kwe-PM10.Izinhlayiya zibambeke ezihlungini ezino-99.9%.ukugcinwa kwe-aerosol, igcinwe kahle futhi ilethwe elabhorethri ye-Applied and Comparative Genomics yase-Trieste University.Uma kubhekwa imvelo "yemvelo" yesampula, okungenzeka ukuthi inothile ngama-inhibitors e-DNA polymerase, saqhubeka nokukhipha i-RNA ngokusebenzisa ikhithi ye-Quick RNA fecal inhlabathi encane eguqulelwe ngohlobo lwezihlungi.[18]Isihlungi sasigoqiwe, uhlangothi olungaphezulu lubheke ngaphakathi,eshubhuni ye-polypropylene engu-5 ml, kanye nobuhlalu obunikezwe kukhithi.Kusukela ku-1 ml wokuqala we-lysisbuffer, sikwazile ukuthola isisombululo esingaba ngu-400 ul, esabe sesicutshungulwa njengoba kuchazwe yizimiso ezijwayelekile, okuholele ekuphumeni kokugcina kwe-15 ul.Kamuva, kwasetshenziswa ama-5 ul ekuhlolweni kwe-SARS-CoV-2.Uma kubhekwa umsuka othile wesampula, kusetshenziswe i-qScript XLT 1-Step RT-qPCR ToughMix.[19]Izinhlelo zokukhuliswa kwakuyilezo zephrothokholi eyakhiwe ngu-Corman et al, eshicilelwe kuwebhusayithi ye-WHO [20].
Ukuhlolwa bekuhloswe ngokusobala ukuqinisekisa noma ukungafaki ubukhona be-SARS-CoV-2 RNA odabeni oluthile.Ukuhlaziywa kokuqala kusebenzise "i-E gene" njengomaka wamangqamuzana futhi kwakhiqiza umphumela omuhle ohlaba umxhwele kuzihlungi eziyi-15 kweziyi-16 ngisho noma, njengoba singalindela, i-Ct yayiphakathi kwemijikelezo engu-36-38.
Ngemuva kwalokho, siphindaphinde ukuhlaziywa kwezihlungi ezi-6 ezinhle (sezivele zilungile ku-“E gene”) ngokusebenzisa i-“RtDR gene” njengomaka wamangqamuzana - okucace kakhulu kwa-SARS-CoV-2 - ukufinyelela emiphumeleni ebalulekile emi-5. of positivity;izivivinyo zokulawula ukukhipha ukuvuma okungamanga nazo zenziwe ngempumelelo (Fig. 1).
Ukuze kugwenywe ukuphela kwezinto zokusampula ezitholakalayo, ama-RNA akhishiwe asele alethwa esibhedlela seNyuvesi yendawo (esinye sezikhungo zomtholampilo ezigunyazwe uHulumeni wase-Italy zokwenza izivivinyo zokuxilonga ze-SARS-CoV-2), ukuze kwenziwe umzuzwana. parallel blind test.Le labhorethri yesibili yomtholampilo ihlole ukukhishwa kwe-RNA kwe-34 yezakhi zofuzo ze-E, N kanye ne-RdRP, ibike imiphumela emihle engu-7 okungenani yezakhi zofuzo zomaka ezintathu, ezine-positivity eqinisekisiwe ngokwehlukana kuzo zonke izimpawu ezintathu (Fig. 2).Ngenxa yohlobo lwesampula, futhi uma kubhekwa ukuthi ukusampula akuzange kwenzelwe izinjongo zokuxilonga emtholampilo kodwa kwenziwe ukuhlolwa kokungcoliswa kwemvelo (kucatshangelwa nokuthi izihlungi zagcinwa okungenani amasonto amane ngaphambi kokuhlaziywa kofuzo lwamangqamuzana, njengobaumphumela wokuvalwa kwe-Italian), singaqinisekisa ukuthi sikubonise ngokunengqondo ukuba khona kwe-SARS-CoV-2 yegciwane le-RNA ngokuthola “ufuzo lwe-RtDR” olucaciswe kakhulu kuzihlungi eziyi-8.Kodwa-ke, ngenxa yokuntuleka kwezinto ezengeziwe ezivela kuzihlungi, asikwazanga ukuphinda inani elanele lokuhlola ukuze sibonise ukuqiniseka kwawo wonke ama-molecular marker angu-3 ngesikhathi esisodwa.
Lobu ubufakazi bokuqala bokuthi i-SARS-CoV-2 RNA ingaba khona odabeni lwezinhlayiya zangaphandle, ngaleyo ndlela iphakamisa ukuthi, ezimeni zokusimama komoya kanye nokugxila okuphezulu kwe-PM, i-SARS-CoV-2 ingakha amaqoqo ane-PM yangaphandle futhi - ukunciphisa i-coefficient yabo yokusabalalisa - ukuthuthukisa ukuphikelela kwegciwane emkhathini.Ezinye iziqinisekiso zalokhu kwandulelaubufakazi buyaqhubeka, futhi kufanele bufake ukuhlolwa kwesikhathi sangempela mayelana nobungqabavu be-SARS-CoV-2 kanye nobubi bayo lapho ikhangiswe odabeni oluthile.Njengamanje, akukho mibono engenziwa mayelana nokuhlobana phakathi kokuba khona kwaleli gciwane ku-PM kanye nokuqubuka kokuqubuka kwe-COVID-19.Ezinye izindaba okufanele zidingidwe ngokuqondile ukugxila okumaphakathi kwe-PM ekugcineniedingekayo ukuze kube "nomphumela okhuthazayo" wokutheleleka (uma kwenzeka kuqinisekiswa ukuthi i-PM ingase isebenze "njengesithwali" se-viral droplet nuclei), noma ngisho nethuba lethiyori lokugoma ngenxa yokuchayeka komthamo omncane emikhawulweni ephansi ye-PM. .
Fig.1 Amajika okukhulisa izakhi zofuzo E (A) kanye ne-RdRP (B): imigqa eluhlaza imele izihlungi ezihloliwe;nqamula imigqaimele izihlungi zenkomba;imigqa ebomvu imelela ukukhuliswa kwamasampuli amahle.
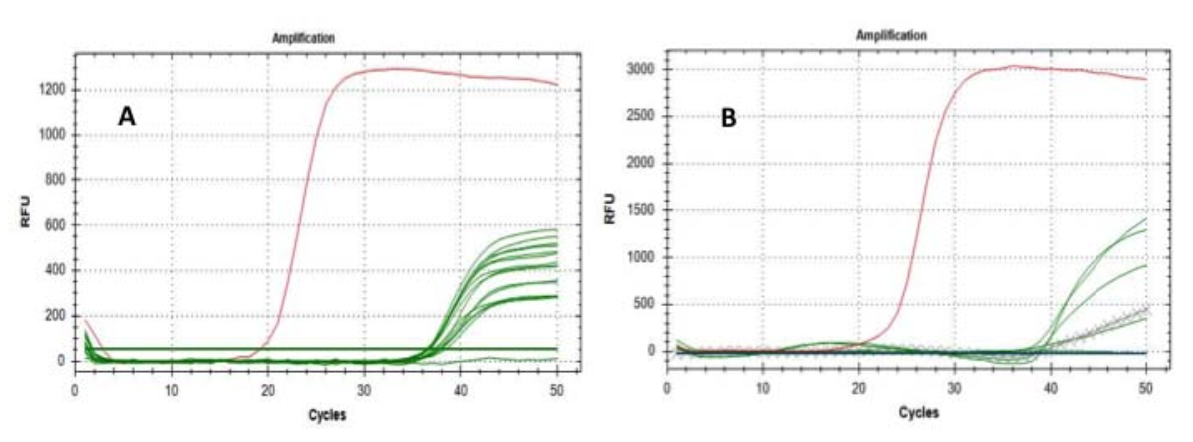
Fig.2.Imiphumela emihle (imakwe ngo-X) yofuzo E, N kanye ne-RdRP etholwe kuwo wonke amasampula angu-34 PM10izihlungi ezihlolwe ekuhlaziyweni okuhambisanayo kwesibili.
 Leonardo Setti1, Fabrizio Passarini2, Gianluigi De Gennaro3, Pierluigi Barbieri4, Maria Grazia Perrone5, Massimo Borelli6, Jolanda Palmisani3, Alessia Di Gilio3, Valentina Torboli6, Alberto Pallavicini6, Maurizio Ruscio7, Prisco Pisciro Mi88, Alessia Di Gilio
Leonardo Setti1, Fabrizio Passarini2, Gianluigi De Gennaro3, Pierluigi Barbieri4, Maria Grazia Perrone5, Massimo Borelli6, Jolanda Palmisani3, Alessia Di Gilio3, Valentina Torboli6, Alberto Pallavicini6, Maurizio Ruscio7, Prisco Pisciro Mi88, Alessia Di Gilio
1. Umnyango Wekhemistry Yezimboni, Inyuvesi yase-Bologna, i-Viale del Risorgimento - 4, I-40136, Bologna, Italy
e-mail: leonardo.setti@unibo.it
2. Isikhungo Seminyango Ephakathi Socwaningo Lwezimboni “Imithombo Evuselelekayo, Imvelo, Ukukhula Okuluhlaza Okuluhlaza, Amandla”,
University of Bologna, Rimini, Italy e-mail: fabrizio.passarini@unibo.it
3. UMnyango Wezemvelo, Inyuvesi “Aldo Moro” yaseBari, Bari, Italy
e-mail: gianluigi.degennaro@uniba.it; alessia.digilio@uniba.it; jolanda.palmisani@uniba.it
4. Umnyango Wesayensi Yamakhemikhali Nezemithi, iNyuvesi yaseTrieste, e-Trieste, e-Italy
e-mail: barbierp@units.it
5. I-Environmental Research Division, i-TCR TECORA, iMilan, e-Italy
e-mail: mariagrazia.perrone@tcrtecora.com
6. UMnyango Wezesayensi Yezempilo - Inyuvesi yaseTrieste, e-Trieste, e-Italy
e-mail: borelli@units.it; torboli@units.it; pallavic@units.it
7. Uphiko Lwezokwelapha Zaselabhorethri, Isibhedlela Senyuvesi Giuliano Isontina (ASU GI), Trieste, Italy
email: maurizio.ruscio@asugi.sanita.fvg.it
8. I-Italian Society of Environmental Medicine (SIMA), iMilan, e-Italy
e-mail: priscofreedom@hotmail.com; alessandro.miani@unimi.it
9. UMnyango Wezesayensi Yezemvelo kanye nePoicy, iNyuvesi yaseMilan, eMilan, e-Italy
e-mail: priscofreedom@hotmail.com; alessandro.miani@unimi.it
Umbhali ohambelanayo:
Leonardo Setti, Department of Industrial Chemistry, University of Bologna Viale del Risorgimento 4, 40136, Bologna, Italy; e-mail: leonardo.setti@unibo.it
Izikhombo
1. Inhlangano Yezempilo Yomhlaba, Izindlela Zokudlulisela Igciwane elibangela i-COVID-19: imithelela yezincomo zokuqapha ze-IPC, Kafushane ngezesayensi;itholakala ku: https://www.who.int/newsroom/commentaries/detail/modes-of-transmission-of-virus-causing-covid-19-implications-for-ipcprecaution-recommendations (29 March 2020)
2. Umnyango Wezempilo wase-Italy, umbiko wansuku zonke wokuqubuka kwe-Covid-19 e-Italy, utholakala ku-http://www.salute.gov.it/imgs/C_17_notizie_4451_0_file.pdf
3. I-EEA, i-European Environmental Agency, Umbiko Wekhwalithi Yomoya e-Europe 2019;Cha 10/2019;I-European Environment Agency: Copenhagen, Denmark, iyatholakala ku: https://www.eea.europa.eu/publications/airquality-in-europe-2019
4. U-Xiao Wu, u-Rachel C. Nethery, M. Benjamin Sabath, u-Danielle Braun, u-Francesca Dominici, Ukuchayeka ekungcoleni komoya kanye nokufa kwe-COVID-19 e-United States, kutholakala kokuthi: https://projects.iq.harvard.edu/ amafayela/covid-pm/files/pm_and_covid_mortality.pdf
5. I-Italian Society of Environmental Medicine (SIMA), Position Paper Particulate Matter kanye ne-COVID-19,
itholakala ku-: http://www.simonlus.it/wpsima/wp-content/uploads/2020/03/COVID_19_positionpaper_ENG.pdf
6. Setti L., Passarini F., De Gennaro G., Barbieri P., Perrone MG, Piazzalunga A., Borelli M., Palmisani J., Di Gilio A, Piscitelli P, Miani A., Is there a Plausible Role nge-Particulate Matter ekusakazeni kwe-COVID-19 eNyakatho ne-Italy?, Izimpendulo Ezisheshayo ze-BMJ, Ephreli 8, 2020, zitholakala ku-: https://www.bmj.com/content/368/bmj.m1103/rapid-responses
7. USedlmaier, N., Hoppenheidt, K., Krist, H., Lehmann, S., Lang, H., Buttner, M. Generation of avian influenza virus (AIV) contaminated fecal fine particulate matter (PM2.5): ukutholwa kwe-genome kanye nokutheleleka kanye nokubalwa kokungena.I-Veterinary Microbiology.139, 156-164 (2009)
8. U-Zhao, Y., Richardson, B., Takle, E., Chai, L., Schmitt, D., Win, H. Ukudluliselwa kwe-Airborne kungenzeka kube neqhaza ekusakazekeni kwe-2015 ukuqubuka komkhuhlane wezinyoni wezinyoni Iziwe Ezihlangene.I-Sci Rep. 9, 11755. https://doi.org/10.1038/s41598-019-47788-z (2019)
9. Ma, Y., Zhou, J., Yang, S., Zhao, Y., Zheng, X. Ukuhlolwa komthelela wezenzakalo zothuli ezenzweni zesimungumungwane entshonalanga ye-China.I-Atmospheric Environment.157, 1-9 (2017)
10. I-Sorensen, JH, Mackay, DKJ, Jensen, C. Ø., Donaldson, AI Imodeli ehlanganisiwe yokubikezela ukusabalala komkhathi kwegciwane lesifo sonyawo nomlomo i-Epidemiol.Infect., 124, 577–590 (2000)
11. Glostera, J., Alexandersen, S. Izikhombisi-ndlela Ezintsha: Ukudluliselwa Emoyeni Kwegciwane Lesifo Sonyawo Nomlomo, 38 (3), 503-505 (2004)
12. Reche, I., D'Orta, G., Mladenov, N., Winget, DM, Suttle, CA Amazinga okubekwa kwamagciwane namagciwane ngaphezu kongqimba lomngcele womkhathi.Ijenali ye-ISME.12, 1154-1162 (2018)
13. Qin, N., Liang, P., Wu, C., Wang, G., Xu, Q., Xiong, X., Wang, T., Zolfo, M., Segata, N., Qin, H ., Knight, R., Gilbert, JA, Zhu, TF Longitudinal survey ye-microbiome ehlotshaniswa ne-particle matter ku-megacity.I-Genome Biology.21, 55 (2020)
14. U-Zhao, Y., Richardson, B., Takle, E., Chai, L., Schmitt, D., Win, H. Ukudluliselwa kwe-Airborne kungase kube
yabamba iqhaza ekubhebhethekeni komkhuhlane wezinyoni wezinyoni obangela izifo ngo-2015 e-United States.Isayensi
I-Rep. 9, 11755. https://doi.org/10.1038/s41598-019-47788-z (2019)
15. Ma, Y., Zhou, J., Yang, S., Zhao, Y., Zheng, X. Ukuhlolwa komthelela wezenzakalo zothuli ezenzweni zesimungumungwane entshonalanga ye-China.I-Atmospheric Environment.157, 1-9 (2017)
16. Jiang, W., Laing, P., Wang, B., Fang, J.,Lang, J., Tian, G., Jiang, J., Zhu, TF Ukukhishwa kwe-DNA Okulungiselelwe kanye nokulandelana kwe-metagenomic kwemiphakathi etholakala emoyeni ye-microbial .Nat.I-Protoc.10, 768-779 (2015)
17. I-Pan, M., Lednicky, JA, Wu, C.-Y., Iqoqo, ukulinganiswa kwezinhlayiyana nokutholwa kwamagciwane asemoyeni.Ijenali ye-Applied Microbiology, 127, 1596-1611 (2019)
18. I-Zymoresearch Ldt, incazelo yomkhiqizo, itholakala ku-: https://www.zymoresearch.com/products/quick-rnafecal-soil-microbe microprep-kit
19. I-Quantabio Ltd, incazelo yomkhiqizo, itholakala ku-: https://www.quantabio.com/qscript-xlt-1-steprt-qpcr-toughmix
20. Corman, VM, Landt, O., Kaiser, M., Molenkamp, R., Meijer, A., Chu, DK, & Mulders, DG (2020).
Ukutholwa kwe-coronavirus yenoveli ka-2019 (2019-nCoV) nge-RT-PCR yesikhathi sangempela.I-Eurosurveillance, 25(3), itholakala ku-:.https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6988269/
Umthombo: https://doi.org/10.1101/2020.04.15.20065995
Isikhathi sokuthumela: Apr-18-2020
